Visualization of Results from many Experiments
Apart from troubleshooting, visualizing the results of a single experiment is of limited use. In this section, we show how to create comparative plots, using results of many experiment. We will use results from the study detailed above.
A First Comparative Plot
Here is the code for generating result plots for two of the benchmarks:
from typing import Dict, Any, Optional
import logging
from baselines import methods
from benchmark_definitions import benchmark_definitions
from syne_tune.experiments import ComparativeResults, PlotParameters, SubplotParameters
def metadata_to_setup(metadata: Dict[str, Any]) -> Optional[str]:
# The setup is the algorithm. No filtering
return metadata["algorithm"]
SETUPS_RIGHT = ("ASHA", "SYNCHB", "BOHB")
def metadata_to_subplot(metadata: Dict[str, Any]) -> Optional[int]:
return int(metadata["algorithm"] in SETUPS_RIGHT)
if __name__ == "__main__":
logging.getLogger().setLevel(logging.INFO)
experiment_name = "docs-1"
experiment_names = (experiment_name,)
setups = list(methods.keys())
num_runs = 15
download_from_s3 = False # Set ``True`` in order to download files from S3
# Plot parameters across all benchmarks
plot_params = PlotParameters(
xlabel="wall-clock time",
aggregate_mode="iqm_bootstrap",
grid=True,
)
# We would like two subplots (1 row, 2 columns), with MOBSTER and HYPERTUNE
# results on the left, and the remaining baselines on the right. Each
# column gets its own title, and legends are shown in both
plot_params.subplots = SubplotParameters(
nrows=1,
ncols=2,
kwargs=dict(sharey="all"),
titles=["Model-based Methods", "Baselines"],
legend_no=[0, 1],
)
# The creation of ``results`` downloads files from S3 (only if
# ``download_from_s3 == True``), reads the metadata and creates an inverse
# index. If any result files are missing, or there are too many of them,
# warning messages are printed
results = ComparativeResults(
experiment_names=experiment_names,
setups=setups,
num_runs=num_runs,
metadata_to_setup=metadata_to_setup,
plot_params=plot_params,
metadata_to_subplot=metadata_to_subplot,
download_from_s3=download_from_s3,
)
# We can now create plots for the different benchmarks
# First: nas201-cifar100
benchmark_name = "nas201-cifar100"
benchmark = benchmark_definitions[benchmark_name]
# These parameters overwrite those given at construction
plot_params = PlotParameters(
metric=benchmark.metric,
mode=benchmark.mode,
ylim=(0.265, 0.31),
)
results.plot(
benchmark_name=benchmark_name,
plot_params=plot_params,
file_name=f"./{experiment_name}-{benchmark_name}.png",
)
# Next: nas201-ImageNet16-120
benchmark_name = "nas201-ImageNet16-120"
benchmark = benchmark_definitions[benchmark_name]
# These parameters overwrite those given at construction
plot_params = PlotParameters(
metric=benchmark.metric,
mode=benchmark.mode,
ylim=(0.535, 0.58),
)
results.plot(
benchmark_name=benchmark_name,
plot_params=plot_params,
file_name=f"./{experiment_name}-{benchmark_name}.png",
)
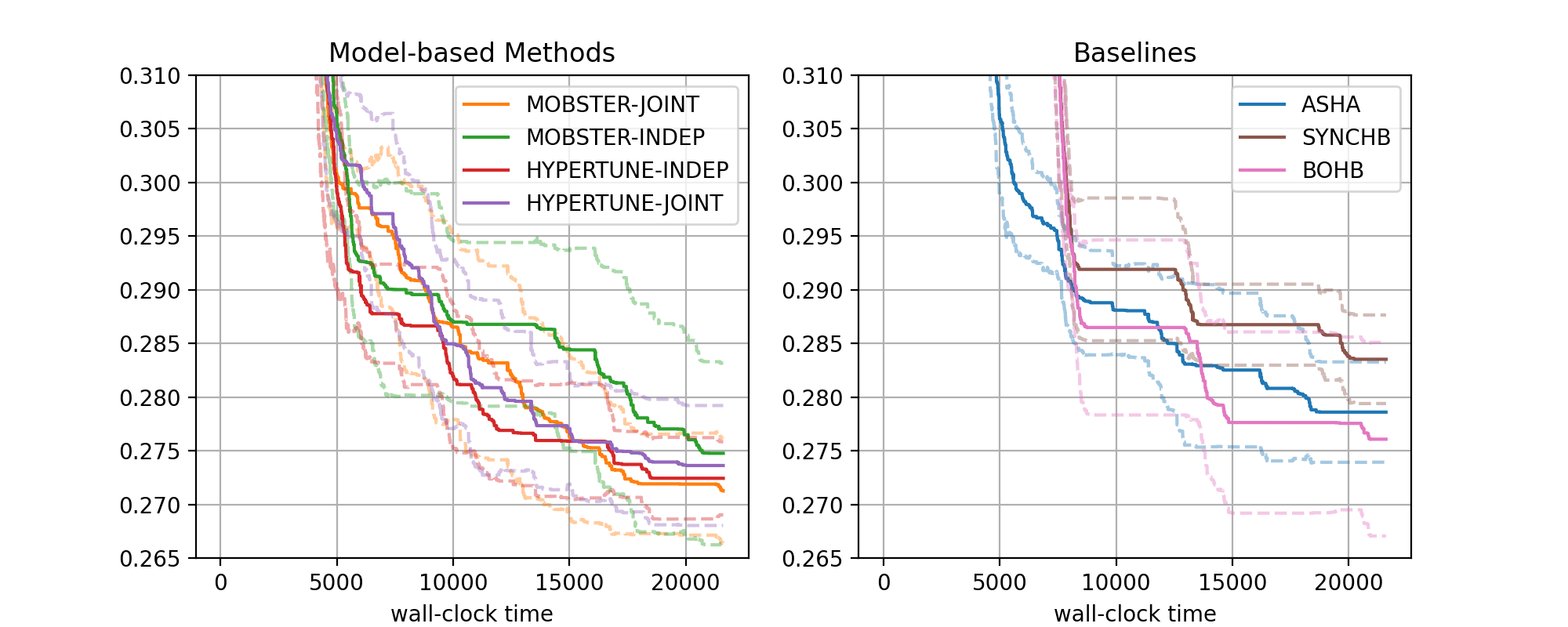
The figure for benchmark nas201-cifar-100 looks as follows:
|
|---|
Results for NASBench-201 (CIFAR-100) |
There are two subfigures next to each other. Each contains a number of curves in bold, along with confidence intervals. The horizontal axis depicts wall-clock time, and on the vertical axis, we show the best metric value found until this time.
More general, the data from our 1260 experiments can be grouped w.r.t. subplot, then setup. Each setup gives rise to one curve (bold, with confidence band). Subplots are optional, the default is to plot a single figure.
The function
metadata_to_setupmaps the metadata stored for an experiment to the setup name, or toNoneif this experiment should be filtered out. In our basic case, the setup is simply the name of the tuning algorithm. Our experimentation framework stores a host of information as metadata, the most useful keys for grouping are:algorithm: Name of method (ASHA,MOBSTER-INDEP, … in our example)tag: Experiment tag. This isdocs-1in our example. Becomes useful when we merge data from different studies in a single figurebenchmark: Benchmark name (nas201-cifar-100, … in our example)n_workers: Number of workers
Other keys may be specific to
algorithm.Once the data is grouped w.r.t. benchmark, then subplot (optional), then setup, we should be left with 15 experiments, one for each seed. Each seed gives rise to a best metric value curve. A metric value
metric_valis converted asmetric_multiplier * metric_valifmode == "min", and as1 - metric_multiplier * metric_valifmode == "max". For example, if your metric is accuracy in percent (from 0 to 100), thenmode="max"andmetric_multiplier=0.01, and the curve shows error in [0, 1].These 15 curves are now interpolated to a common grid, and at each grid point, the 15 values (one for each seed) are aggregated into 3 values
lower,aggregate,upper. In the figure,aggregateis shown in bold, andlower,upperin dashed. Different aggregation modes are supported (selected byplot_params.aggregate_mode):mean_and_ci: Mean and 0.95 normal confidence intervaliqm_bootstrap(default): Interquartile mean and 0.95 confidence interval based on the bootstrap variance estimate. These statistics are argued for in Agarwal et.al: Deep Reinforcement Learning at the Edge of the Statistical Precipice.median_percentiles: Median and 25 (lower), 75 (upper) percentiles
Plotting starts with the creation of a
ComparativeResultsobject. We need to pass the experiment names (or tags), the list of all setups, the number of runs (or seeds), themetadata_to_setupfunction, as well as default plot parameters inplot_params. SeePlotParametersfor full details about the latter. In our example, we setxlabel,aggregate_mode(see above), and enable a grid withgrid=True. Note that these parameters can be extended and overwritten by parameters for each plot.In our example, we separate the MOBSTER and HYPERTUNE setups from the baselines, by using two subfigures. This is done by specifying
plot_params.subplotsandmetadata_to_subplot. In the former,plot_params.subplots.nrowsandplot_params.subplots.ncols `` are mandatory, prescribing the shape of the subplot arrangement. In ``plot_params.subplots.titles, we can provide titles for each column (which we do here). If given, this overridesplot_params.title. Also,plot_params.subplots.legend_no=[0, 1]asks for legends in both subplots (the default is no legend at all). For full details about these arguments, seeSubplotParametersThe creation of
resultsdoes a number of things. First, ifdownload_from_s3=True, result files are downloaded from S3. In our example, we assume this has already been done. Next, all result files are iterated over, allmetadata.jsonare read, and an inverse index from benchmark name to paths,setup_name, andsubplot_nois created. This process also checks that exactlynum_runsexperiments are present for every setup. For large studies, it frequently happens that too few or too many results are found. The warning outputs can be used for debugging.Given
results, we can create plots for every benchmark. In our example, this is done fornas201-cifar100andnas201-ImageNet16-120, by callingresults.plot(). Apart from the benchmark name, we also pass plot parameters inplot_params, which extend (and overwrite) those passed at construction. In particular, we need to passmetricandmode, which we can obtain from the benchmark description. Moreover,ylimis a sensible range for the vertical axis, which is different for every benchmark (this is optional).If we pass
file_nameas argument toresults.plot, the figure is stored in this file.results.plotreturns a dictionary, whose entries “fig” and “axs” contain the figure and its axes (subfigures), allowing for further fine-tuning.
Note
If suplots are used, the grouping is w.r.t. (subplot, setup), not
just by setup. This means you can use the same setup name in
different subplots to show different data. For example, your study may
have run a range of methods under different conditions (say, using a
different number of workers). You can then map these conditions to
subplots and show the same setups in each subplot. In any case, the
mapping of setups to colors is fixed and the same in every subplot.
Note
Plotting features presented here can also be used to visualize results for a single seed. In this case, there are no error bars.
Additional Features
In this section, we discuss additional features, allowing you to customize your result plots.
Combining Results from Multiple Studies
HPO experiments are expensive to do, so you want to avoid re-running them for baselines over and over. Our plotting tools allow you to easily combine results across multiple studies.
As an example, say we would like to relate our docs-1 results to what
random search and Bayesian optimization do on the same benchmarks. These
baseline results were already obtained as part of an earlier study
baselines-1, in which a number of methods were compared, among them RS
and BO. As an additional complication, the earlier study used 30
repetitions (or seeds), while docs-1 uses 15. Here is the modification of
the code above in order to include these additional baseline results in the
plot on the right side. First, we need to replace metadata_to_setup and
SETUPS_RIGHT:
def metadata_to_setup(metadata: Dict[str, Any]) -> Optional[str]:
algorithm = metadata["algorithm"]
tag = metadata["tag"]
seed = int(metadata["seed"])
# Filter out experiments from "baselines-1" we don't want to compare
# against
if tag == "baselines-1" and (seed >= 15 or algorithm not in ("RS", "BO")):
return None
else:
return algorithm
SETUPS_RIGHT = ("ASHA", "SYNCHB", "BOHB", "RS", "BO")
There are now two more setups, “RS” and “BO”, whose results come from the
earlier baselines-1 study. Now, ComparativeResults has to be created
differently:
experiment_names = experiment_names + ("baselines-1",)
setups = setups + ["RS", "BO"]
results = ComparativeResults(
experiment_names=experiment_names,
setups=setups,
num_runs=num_runs,
metadata_to_setup=metadata_to_setup,
plot_params=plot_params,
metadata_to_subplot=metadata_to_subplot,
download_from_s3=download_from_s3,
)
Note
If you intend to combine results from several different studies, it is
recommended to use the same random seed (specified as --random_seed),
which ensures that the same sequence of random numbers is used in each
experiment. This results in a so-called paired comparison, lowering the
random variations across setups. In our example, we would look up the
master random seed of the baselines-1 study and use this for docs-1
as well.
Add Performance of Initial Trials
When using HPO, you often have an idea about one or several default
configurations that should be tried first. In Syne Tune, such initial
configurations can be specified by points_to_evaluate (see
here for details).
An obvious question to ask is how long it takes for a HPO method to find a
configuration which works significantly better than these initial ones.
We can visualize the performance of initial trials by specifying
plot_params.show_init_trials of type
ShowTrialParameters. In our docs-1 study,
points_to_evaluate is not explicitly used, but the configuration of the
first trial is selected by a mid-point heuristic. Our plotting script from
above needs to be modified:
plot_params.show_init_trials = ShowTrialParameters(
setup_name="ASHA",
trial_id=0,
new_setup_name="default"
)
results = ComparativeResults(
experiment_names=experiment_names,
setups=setups,
num_runs=num_runs,
metadata_to_setup=metadata_to_setup,
plot_params=plot_params,
metadata_to_subplot=metadata_to_subplot,
download_from_s3=download_from_s3,
)
Since the ASHA curve is plotted on the right side, this will add another
curve there with label default. This curve shows the best metric value,
using data from the first trial only (trial_id == 0). It is extended as a
flat constant line to the end of the horizontal range.
If you specify a number of initial configurations with points_to_evaluate,
set ShowTrialParameters.trial_id to their number minus 1. The initial trials
curve will use data from trials with ID less or equal this number.
Controlling Subplots
Our example above already creates two subplots, horizontally arranged, and we
discussed the role of metadata_to_subplot. Here, we provide extra details
about fields in SubplotParameters, the type
for plot_params.subplots:
nrows,ncols: Shape of subplot matrix. The total number of subplots is<= ncols * nrows.kwargscontains further arguments tomatplotlib.pyplot.subplots. For example, ifsharey="all", the y tick labels are only created for the first column. If you usenrows > 1, you may want to share x tick labels as well, withsharex="all".titles: Iftitle_each_figure == False, this is a list of titles, one for each column. Iftitle_each_figure == True, thentitlescontains a title for each subplot. Iftitlesis not given, the global titleplot_params.titleis printed on top of the left-most column.legend_no: List of subfigures in which the legend is shown. The default is not to show legends. In our example, there are different setups in each subplot, so we want a legend in each. If your subplots show the same setups under different conditions, you may want to show the legend in one of the subplots only, in which caselegend_nocontains a single number.xlims: Use this if your subfigures have x axis ranges. The globalxlimis overwritten by(0, xlims[subplot_no]).subplot_indices: Any given plot produced byplot()does not have to contain all subfigures. For example, you may want to group your results into 4 or 8 bins, then create a sequence of plots comparing pairs of them. Ifsubplot_indicesis given, it contains the subplot indices to be shown, and this order. Otherwise, this is \(0, 1, 2, \dots\). If this is given, thentitlesandxlimsis relative to this list (in thatxlims[i]corresponds to subfiguresubplot_indices[i]), butlegend_nois not.
Plotting Derived Metrics
You can also plot metrics which are not directly contained in the results data
(as a column), but which can be computed from the results. To this end, you
can pass a dataframe column generator as dataframe_column_generator to
plot(). For example, assume
we run multi-objective HPO methods on a benchmark involving metrics cost
and latency (mode="min" for both of them). The final plot command
would look like this:
from syne_tune.experiments.multiobjective import (
hypervolume_indicator_column_generator,
)
# ...
dataframe_column_generator = hypervolume_indicator_column_generator(
metrics_and_modes = [("cost", "min"), ("latency", "min")]
)
plot_params = PlotParameters(
metric="hypervolume_indicator",
mode="max",
)
results.plot(
benchmark_name=benchmark_name,
plot_params=plot_params,
dataframe_column_generator=dataframe_column_generator,
one_result_per_trial=True,
)
The mapping returned by
hypervolume_indicator_column_generator()maps a results dataframe to a new column containing the best hypervolume indicator as function of wall-clock time for the metricscostandlatency, which must be contained in the results dataframe.The option
one_result_per_trial=Trueofresults.plotensures that the result data is filtered, so that for each experiment, one the final row for each trial remains. This option is useful if the methods are single-fidelity, but results are reported after each epoch. The filtering makes sure that only results for the largest epoch are used for each trial. Since this is done before the best hypervolume indicator is computed, it can speed up the computation dramatically.
Filtering Experiments by DateTime Bounds
Results can be filtered out by having metadata_to_setup or
metadata_to_subplot return None. This is particularly useful if results
from several studies are to be combined. Another way to filter experiments is
using the datetime_bounds argument of
ComparativeResults. A common use case is that
experiments for a large study have been launched in several stages, and those
of an early stage failed. If the corresponding result files are not removed on S3,
the creation of ComparativeResults will complain about too many results
being found. datetime_bounds is specified in terms of date-time strings of
the format ST_DATETIME_FORMAT, which currently is
“YYYY-MM-DD-HH-MM-SS”. For example, if results are valid from
“2023-03-19-22-01-57” onwards, but invalid before, we can use
datetime_bounds=("2023-03-19-22-01-57", None). datetime_bounds can also
be a dictionary with keys from experiment_names, in which case bounds are
specific to different experiment prefixes.
Extract Meta-Data Values
Apart from plotting results, we can also retrieve meta-data values. This is
done by passing a list of meta-data key names via metadata_keys when
creating ComparativeResults. Afterwards, the
corresponding meta-data values can be queried by calling
results.metadata_values(benchmark_name). The result is a nested dictionary
result, so that result[key][setup_name] is a list of values, where
key is the meta-data key from metadata_keys, setup_name is a setup
name. The list contains values from all experiments mapped to this
setup_name. If you use the same setup names across different subplots,
set metadata_subplot_level=True, in which case
results.metadata_values(benchmark_name) returns
result[key][setup_name][subplot_no], so the grouping w.r.t. setup names
and subplots is used.
Extract Final Values for Extra Results
Syne Tune allows extra results to be stored alongside the usual metrics data,
as shown in
examples/launch_height_extra_results.py.
These are simply additional columns in the result dataframe. In order to plot
them over time, you currently need to write your own plotting scripts. If the
best value over time approach of Syne Tune’s plotting tools makes sense for
any single column, you can just specify their name for plot_params.metric
and set plot_params.mode accordingly.
However, in many cases it is sufficient to know final values for extra results,
grouped in the same way as everything else. For example, extra results may be
used to monitor some internals of the HPO method being used, in which case we
may be satisfied to see these statistics at the end of experiments. If
extra_results_keys is used in
plot(), the method returns
a nested dictionary extra_results under key “extra_results”, so that
extra_results[setup_name][key] contains a list of values (one for each
seed) for setup setup_name and key an extra result name from
extra_results_keys. As above, if
metadata_subplot_level=True at construction of
ComparativeResults, the structure of the
dictionary is extra_results[setup_name][subplot_no][key].